Mixture Model¶
This notebook contains the code for a simple implementation of the Leaspy Mixture model on synthetic data. Before implementing the model take a look at the relevant mathematical framework in the user guide.
The following imports are required libraries for numerical computation, data manipulation, and visualization.
import os
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import torch
import leaspy
from leaspy.io.data import Data
This toy example is part of a simulation study, carried out by Sofia Kaisaridi that will be included in an article to be submitted in a biostatistics journal. The dataset contains 1000 individuals each with 6 visits and 6 scores.
leaspy_root = os.path.dirname(leaspy.__file__)
data_path = os.path.join(leaspy_root, "datasets/data/simulated_data_for_mixture.csv")
all_data = pd.read_csv(data_path, sep=";", decimal=",")
all_data["ID"] = all_data["ID"].ffill()
all_data = all_data.set_index(["ID", "TIME"])
all_data.head()
| score_1_normalized | score_2_normalized | score_3_normalized | score_4_normalized | score_5_normalized | score_6_normalized | ||
|---|---|---|---|---|---|---|---|
| ID | TIME | ||||||
| subj_1 | 50.159062 | 0.197863 | 0.057787 | 0.095305 | 0.141747 | 0.265511 | 0.397614 |
| 66.713371 | 0.418290 | 0.139975 | 0.021792 | 0.119112 | 0.481825 | 0.456491 | |
| 68.994947 | 0.397005 | 0.149633 | 0.040861 | 0.128334 | 0.388164 | 0.394283 | |
| 73.009543 | 0.330076 | 0.127369 | 0.098300 | 0.348438 | 0.473187 | 0.418984 | |
| 80.948301 | 0.530554 | 0.117444 | 0.159612 | 0.339971 | 0.468804 | 0.591484 |
We load the Mixture Model from the leaspy library and transform the dataset in a leaspy-compatible form with the built-in functions.
from leaspy.models import LogisticMultivariateMixtureModel
leaspy_data = Data.from_dataframe(all_data)
Then we fit a model with 3 clusters and 2 sources. Note that we have an extra argument n_clusters than the
standard model that has to be specified in order for the mixture model to run.
model = LogisticMultivariateMixtureModel(
name="multi",
source_dimension=2,
dimension=6,
n_clusters=3,
obs_models="gaussian-diagonal",
)
model.fit(leaspy_data, "mcmc_saem", seed=1312, n_iter=100, progress_bar=False)
model.summary()
Fit with `AlgorithmName.FIT_MCMC_SAEM` took: 3.96s
================================================================================
Model Summary
================================================================================
Model Name: multi
Model Type: LogisticMultivariateMixtureModel
Features (6): score_1_normalized, score_2_normalized,
score_3_normalized, score_4_normalized, score_5_normalized,
score_6_normalized
Sources (2): Source 0 (s0), Source 1 (s1)
Clusters (3): Cluster 0 (c0), Cluster 1 (c1), Cluster 2 (c2)
Observation Models: gaussian-diagonal
Neg. Log-Likelihood: -5265.7297
Training Metadata
--------------------------------------------------------------------------------
Algorithm: AlgorithmName.FIT_MCMC_SAEM
Seed: 1312
Iterations: 100
Data Context
--------------------------------------------------------------------------------
Subjects: 1000
Visits: 6000
Total Observations: 36000
Leaspy Version: 2.0.1
================================================================================
Population Parameters
--------------------------------------------------------------------------------
betas_mean:
s0 s1
b0 -0.0099 0.0054
b1 -0.0062 0.0054
b2 0.0372 -0.0189
b3 0.0329 -0.0143
b4 0.0424 -0.0385
c0 c1 c2
probs 0.3247 0.4001 0.2752
sources_mean:
c0 c1 c2
s0 -1.1791 -0.9338 2.7506
s1 2.4279 -0.8996 -1.5589
score_1. score_2. score_3. score_4. score_5. score_6.
log_g_mean 0.6386 1.6792 2.2111 0.9981 0.4451 -0.0358
score_1. score_2. score_3. score_4. score_5. score_6.
log_v0_mean -5.0169 -5.0958 -6.3110 -5.1638 -5.3070 -5.6924
Individual Parameters
--------------------------------------------------------------------------------
c0 c1 c2
tau_mean 71.0264 66.0683 57.4709
c0 c1 c2
tau_std 10.4716 8.8100 11.6153
c0 c1 c2
xi_mean -0.1970 -0.2768 0.6348
c0 c1 c2
xi_std 0.8801 0.9058 1.0671
Noise Model
--------------------------------------------------------------------------------
score_1. score_2. score_3. score_4. score_5. score_6.
noise_std 0.0744 0.0715 0.0370 0.0625 0.0812 0.0779
================================================================================
First we take a look in the population parameters.
With the mixture model we obtain separate values for the tau_mean, xi_mean and the sources_mean for each cluster,
as well as the cluster probabilities (probs).
print(model.parameters)
{'betas_mean': tensor([[-0.0099, 0.0054],
[-0.0062, 0.0054],
[ 0.0372, -0.0189],
[ 0.0329, -0.0143],
[ 0.0424, -0.0385]]), 'log_g_mean': tensor([ 0.6386, 1.6792, 2.2111, 0.9981, 0.4451, -0.0358]), 'log_v0_mean': tensor([-5.0169, -5.0958, -6.3110, -5.1638, -5.3070, -5.6924]), 'noise_std': tensor([0.0744, 0.0715, 0.0370, 0.0625, 0.0812, 0.0779], dtype=torch.float64), 'probs': tensor([0.3247, 0.4001, 0.2752], dtype=torch.float64), 'sources_mean': tensor([[-1.1791, -0.9338, 2.7506],
[ 2.4279, -0.8996, -1.5589]], dtype=torch.float64), 'tau_mean': tensor([71.0264, 66.0683, 57.4709], dtype=torch.float64), 'tau_std': tensor([10.4716, 8.8100, 11.6153], dtype=torch.float64), 'xi_mean': tensor([-0.1970, -0.2768, 0.6348], dtype=torch.float64), 'xi_std': tensor([0.8801, 0.9058, 1.0671], dtype=torch.float64)}
Then we can also retrieve the individual parameters and the posteriors probabilities of cluster membership. We then consider that the cluster label is the cluster with the biggest probability.
from torch.distributions import Normal
def get_ip(df_leaspy, model):
"""
leaspy_data : the dataframe with the correct indexing
leaspy_mixture : the leaspy object after the fit
"""
ip = pd.DataFrame(df_leaspy.index.get_level_values("ID").unique(), columns=["ID"])
ip[["tau"]] = model.state["tau"]
ip[["xi"]] = model.state["xi"]
ip[["sources_0"]] = model.state["sources"][:, 0].cpu().numpy().reshape(-1, 1)
ip[["sources_1"]] = model.state["sources"][:, 1].cpu().numpy().reshape(-1, 1)
params = model.parameters
probs = params["probs"]
# Number of individuals and clusters
n = len(ip)
k = len(probs)
means = {
"tau": params["tau_mean"], # shape: (2,)
"xi": params["xi_mean"],
"sources_0": params["sources_mean"][0, :],
"sources_1": params["sources_mean"][1, :],
}
stds = {
"tau": params["tau_std"],
"xi": params["xi_std"],
"sources_0": torch.tensor(1),
"sources_1": torch.tensor(1),
}
stds["sources_0"] = stds["sources_0"].repeat(k)
stds["sources_1"] = stds["sources_1"].repeat(k)
values = {
"tau": torch.tensor(ip["tau"].values),
"xi": torch.tensor(ip["xi"].values),
"sources_0": torch.tensor(ip["sources_0"].values),
"sources_1": torch.tensor(ip["sources_1"].values),
}
# Compute log-likelihoods for each variable
log_likelihoods = torch.zeros((n, k))
for var in ["tau", "xi", "sources_0", "sources_1"]:
x = torch.tensor(ip[var].values)
for cluster in range(k):
dist = Normal(means[var][cluster], stds[var][cluster])
log_likelihoods[:, cluster] += dist.log_prob(x)
# Add log-priors
log_priors = torch.log(probs)
log_posteriors = log_likelihoods + log_priors
# Normalize using logsumexp
log_sum = torch.logsumexp(log_posteriors, dim=1, keepdim=True)
responsibilities = torch.exp(log_posteriors - log_sum)
for i in range(responsibilities.shape[1]):
ip[f"prob_cluster_{i}"] = responsibilities[:, i].numpy()
# Automatically find all probability columns
prob_cols = [col for col in ip.columns if col.startswith("prob_cluster_")]
# Assign the most likely cluster
ip["cluster_label"] = ip[prob_cols].values.argmax(axis=1)
return ip
ip = get_ip(all_data, model)
ip.head()
| ID | tau | xi | sources_0 | sources_1 | prob_cluster_0 | prob_cluster_1 | prob_cluster_2 | cluster_label | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | subj_1 | 74.345524 | 0.425971 | -1.081884 | -1.749747 | 0.000256 | 0.999451 | 2.925267e-04 | 1 |
| 1 | subj_2 | 73.913712 | -0.282853 | 0.135766 | -1.325051 | 0.000707 | 0.989080 | 1.021316e-02 | 1 |
| 2 | subj_3 | 75.480702 | -0.588609 | -0.100608 | 2.657032 | 0.997874 | 0.002125 | 5.157013e-07 | 0 |
| 3 | subj_4 | 67.857884 | 0.277853 | 0.234159 | -0.477420 | 0.008153 | 0.974729 | 1.711859e-02 | 1 |
| 4 | subj_5 | 64.239131 | -0.671196 | -0.949378 | -1.022694 | 0.001409 | 0.998407 | 1.842238e-04 | 1 |
We produce the population progression plots. We separate the model parameters for each cluster and we store them to a dedicated item.
from leaspy.io.outputs import IndividualParameters
parameters = model.parameters
number_of_sources = 2
# cluster 0
mean_xi = parameters["xi_mean"].numpy()[0]
mean_tau = parameters["tau_mean"].numpy()[0]
mean_source = parameters["sources_mean"].numpy()[0, 0].tolist()
mean_sources = [mean_source] * number_of_sources
parameters_0 = {"xi": mean_xi, "tau": mean_tau, "sources": mean_sources}
ip_0 = IndividualParameters()
ip_0.add_individual_parameters("average", parameters_0)
# cluster 1
mean_xi = parameters["xi_mean"].numpy()[1]
mean_tau = parameters["tau_mean"].numpy()[1]
mean_source = parameters["sources_mean"].numpy()[0, 1].tolist()
mean_sources = [mean_source] * number_of_sources
parameters_1 = {"xi": mean_xi, "tau": mean_tau, "sources": mean_sources}
ip_1 = IndividualParameters()
ip_1.add_individual_parameters("average", parameters_1)
# cluster 2
mean_xi = parameters["xi_mean"].numpy()[2]
mean_tau = parameters["tau_mean"].numpy()[2]
mean_source = parameters["sources_mean"].numpy()[0, 2].tolist()
number_of_sources = 2
mean_sources = [mean_source] * number_of_sources
parameters_2 = {"xi": mean_xi, "tau": mean_tau, "sources": mean_sources}
ip_2 = IndividualParameters()
ip_2.add_individual_parameters("average", parameters_2)
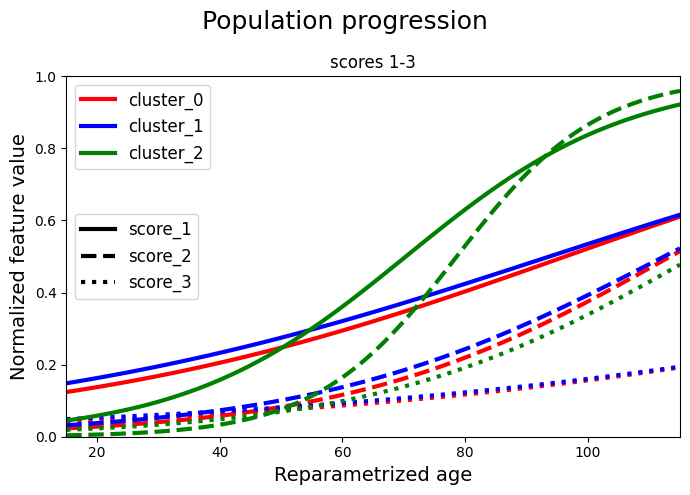
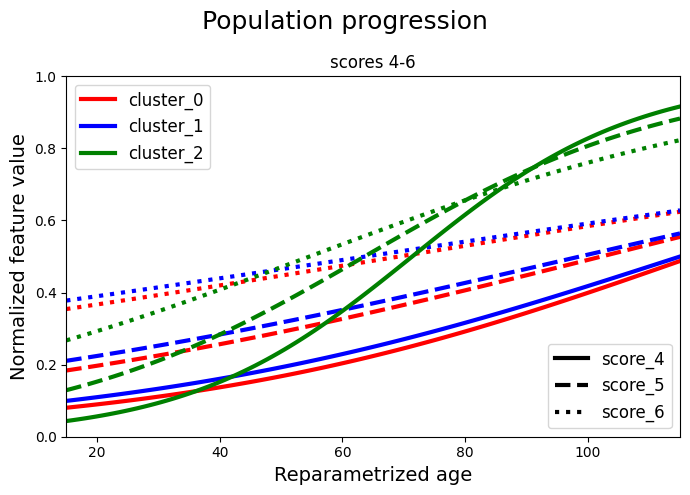
We separate the scores and we plot two graphs for clarity. Each cluster is represented with a different colour. Each score is represented with a different linestyle. In the first graph we have the first 3 scores and in the second graph we have the remaining 3 scores. In each graph we plot all the clusters.
from matplotlib.lines import Line2D
timepoints = np.linspace(15, 115, 105)
values_0 = model.estimate({"average": timepoints}, ip_0)
values_1 = model.estimate({"average": timepoints}, ip_1)
values_2 = model.estimate({"average": timepoints}, ip_2)
# A. Scores 1-3
plt.figure(figsize=(7, 5))
plt.ylim(0, 1)
lines = [
"-",
"--",
":",
]
## cluster 0
for ls, name, val in zip(lines, model.features, values_0["average"][:, 0:3].T):
plt.plot(timepoints, val, label=f"cluster_0_{name}", c="red", linewidth=3, ls=ls)
## cluster 1
for ls, name, val in zip(lines, model.features, values_1["average"][:, 0:3].T):
plt.plot(timepoints, val, label=f"cluster_1_{name}", c="blue", linewidth=3, ls=ls)
## cluster 2
for ls, name, val in zip(lines, model.features, values_2["average"][:, 0:3].T):
plt.plot(timepoints, val, label=f"cluster_2_{name}", c="green", linewidth=3, ls=ls)
## legends
cluster_legend = [
Line2D([0], [0], color="red", linewidth=3, label="cluster_0"),
Line2D([0], [0], color="blue", linewidth=3, label="cluster_1"),
Line2D([0], [0], color="green", linewidth=3, label="cluster_2"),
]
legend1 = plt.legend(handles=cluster_legend, loc="upper left", prop={"size": 12})
color_legend = [
Line2D([0], [0], linestyle=lines[0], color="black", linewidth=3, label="score_1"),
Line2D([0], [0], linestyle=lines[1], color="black", linewidth=3, label="score_2"),
Line2D([0], [0], linestyle=lines[2], color="black", linewidth=3, label="score_3"),
]
legend2 = plt.legend(
handles=color_legend, loc="center left", title_fontsize=14, prop={"size": 12}
)
plt.gca().add_artist(legend1)
plt.xlim(min(timepoints), max(timepoints))
plt.xlabel("Reparametrized age", fontsize=14)
plt.ylabel("Normalized feature value", fontsize=14)
plt.title("scores 1-3")
plt.suptitle("Population progression", fontsize=18)
plt.tight_layout()
plt.show()
# B. Scores 4-6
plt.figure(figsize=(7, 5))
plt.ylim(0, 1)
lines = [
"-",
"--",
":",
]
## cluster 0
for ls, name, val in zip(lines, model.features, values_0["average"][:, 3:6].T):
plt.plot(timepoints, val, label=f"cluster_0_{name}", c="red", linewidth=3, ls=ls)
## cluster 1
for ls, name, val in zip(lines, model.features, values_1["average"][:, 3:6].T):
plt.plot(timepoints, val, label=f"cluster_1_{name}", c="blue", linewidth=3, ls=ls)
## cluster 2
for ls, name, val in zip(lines, model.features, values_2["average"][:, 3:6].T):
plt.plot(timepoints, val, label=f"cluster_2_{name}", c="green", linewidth=3, ls=ls)
## legends
cluster_legend = [
Line2D([0], [0], color="red", linewidth=3, label="cluster_0"),
Line2D([0], [0], color="blue", linewidth=3, label="cluster_1"),
Line2D([0], [0], color="green", linewidth=3, label="cluster_2"),
]
legend1 = plt.legend(handles=cluster_legend, loc="upper left", prop={"size": 12})
color_legend = [
Line2D([0], [0], linestyle=lines[0], color="black", linewidth=3, label="score_4"),
Line2D([0], [0], linestyle=lines[1], color="black", linewidth=3, label="score_5"),
Line2D([0], [0], linestyle=lines[2], color="black", linewidth=3, label="score_6"),
]
legend2 = plt.legend(
handles=color_legend, loc="lower right", title_fontsize=14, prop={"size": 12}
)
plt.gca().add_artist(legend1)
plt.xlim(min(timepoints), max(timepoints))
plt.xlabel("Reparametrized age", fontsize=14)
plt.ylabel("Normalized feature value", fontsize=14)
plt.title("scores 4-6")
plt.suptitle("Population progression", fontsize=18)
plt.tight_layout()
plt.show()
This concludes the Mixture Model example using Leaspy. We can also use these fit models to simulate new data according to the estimated parameters. This can be useful for validating the model, for generating synthetic datasets for further analysis or for generate a trajectory for a new individual given specific parameters. Let’s check this in the next example.