Understanding Leaspy’s Data Containers: Data and Dataset¶
In leaspy, transforming raw data (like a CSV) into a model-ready format involves two key classes: Data and Dataset. Understanding their distinct roles is crucial for having full control of your analysis.
1. The Data Class: The User Interface¶
The Data class is your primary tool for loading, organizing, and inspecting data. It acts as a flexible, patient-centric container that bridges the gap between raw spreadsheets and the model.
Key Features & Methods¶
Loading Data¶
Use the factory method to load from a pandas DataFrame. Notice that there is a slight difference when you work with joint models.
import os
import pandas as pd
import leaspy
from leaspy.io.data import Data
leaspy_root = os.path.dirname(leaspy.__file__)
data_path = os.path.join(leaspy_root, "datasets/data/simulated_data_for_joint.csv")
df = pd.read_csv(data_path, dtype={"ID": str}, sep=";")
data = Data.from_dataframe(df) # <-
# For joint models (longitudinal + time-to-event):
data_joint = Data.from_dataframe(df, data_type='joint') # <-
print(df.head(3))
ID TIME EVENT_TIME EVENT_BOOL Y0 Y1 Y2 Y3
0 116 78.461 85.5 1 0.44444 0.04 0.0 0.0
1 116 78.936 85.5 1 0.60000 0.00 0.0 0.2
2 116 79.482 85.5 1 0.39267 0.04 0.0 0.2
Inspection:¶
Access data naturally by patient ID or index. This is made thanks to the iterators handdling inside Data, it also allows you to iterate using for loops. So you can:
select data usint the brackets (
data['116'])check some attributes (
data.n_individuals)convert the whole dataset or some individuals back into a dataframe object (
data[['116']].to_dataframe())iterate into each individual (
for individual in data:)generate an iterator (
for i, individual in enumerate(data):)
patient_data = data['116'] # Get a specific individual
n_patients = data.n_individuals # Get total count
print(f"Number of patients: {n_patients}")
print(f"patient data (observations shape): {patient_data.observations.shape}")
print(f"patient data (dataframe):\n{data[['116']].to_dataframe()}")
print("\nIterating over first 3 patients:")
for individual in data:
if len(individual.timepoints) == 10: break
print(f" - Patient {individual.idx}: {len(individual.timepoints)} visits")
for i, individual in enumerate(data):
if i >= 3: break # Stop after 3 iterations using the index 'i'
print(f"Patient ID: {individual.idx}")
Number of patients: 17
patient data (observations shape): (9, 6)
patient data (dataframe):
ID TIME EVENT_TIME EVENT_BOOL Y0 Y1 Y2 Y3
0 116 78.461 85.5 1.0 0.44444 0.04 0.0 0.0
1 116 78.936 85.5 1.0 0.60000 0.00 0.0 0.2
2 116 79.482 85.5 1.0 0.39267 0.04 0.0 0.2
3 116 79.939 85.5 1.0 0.58511 0.00 0.0 0.0
4 116 80.491 85.5 1.0 0.57044 0.00 0.0 0.0
5 116 81.455 85.5 1.0 0.55556 0.20 0.1 0.2
6 116 82.491 85.5 1.0 0.71844 0.20 0.1 0.6
7 116 83.463 85.5 1.0 0.71111 0.32 0.2 0.6
8 116 84.439 85.5 1.0 0.91111 0.52 0.6 1.0
Iterating over first 3 patients:
- Patient 116: 9 visits
- Patient 142: 11 visits
- Patient 169: 7 visits
Patient ID: 116
Patient ID: 142
Patient ID: 169
Managing Cofactors¶
Easily attach patient characteristics (e.g., genetics, demographics). It is used to group populations when using plotter.plot_distribution, so plotter can color different cofactors. Lets generate a dataset and its parameters to show how it works.
import numpy as np
import torch
from leaspy.io.logs.visualization import Plotter
from leaspy.io.outputs import Result
n_individuals = 2000
patient_ids = [str(i) for i in range(n_individuals)]
df_longitudinal = pd.DataFrame({'ID': np.repeat(patient_ids, 3), 'TIME': np.tile([60, 70, 80], n_individuals), 'Y0': np.random.rand(n_individuals * 3)})
data = Data.from_dataframe(df_longitudinal)
df_cofactors = pd.DataFrame({'gender': np.random.choice(['Male', 'Female'], size=n_individuals)}, index=patient_ids)
df_cofactors.index.name = 'ID'
data.load_cofactors(df_cofactors, cofactors=['gender'])
individual_parameters = {'tau': torch.tensor(np.random.normal(70, 5, (n_individuals, 1))), 'xi': torch.tensor(np.random.normal(0, 0.5, (n_individuals, 1)))}
result_obj = Result(data, individual_parameters)
Plotter().plot_distribution(result_obj, parameter='tau', cofactor='gender')
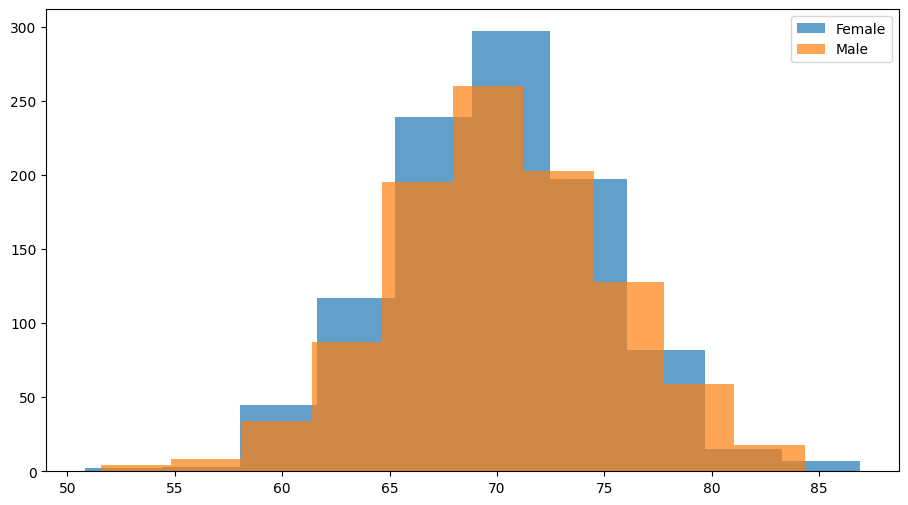
It is important to note that attaching external data to the class through data.load_cofactors is different from loading cofactors inside the model using factory_kws:
Feature |
Covariates (via |
Cofactors (via |
|---|---|---|
Purpose |
Used inside the model to modulate parameters (e.g., in |
Used for analysis/metadata (e.g., plotting, stratification) but ignored by the model’s math. |
Loading |
Loaded during |
Loaded after |
Storage |
Stored as a |
Stored as a |
Constraints |
Must be integers, constant per individual, and have no missing values. |
Can be any type (strings, floats, etc.). |
Best Practice:¶
Always create a Data object first. It validates your input and handles irregularities (missing visits, different timelines) gracefully.
2. The Dataset Class: The Internal Engine¶
The Dataset class is the high-performance numerical engine. It converts the flexible Data object into rigid PyTorch Tensors optimized for mathematical computation.
What it does¶
Tensorization: Converts all values to PyTorch tensors.
Padding: Standardizes patient timelines by padding them to the maximum number of visits (creating a rectangular data block).
Masking: Creates a binary mask to distinguish real data from padding.
When to use it explicitly?¶
You rarely need to instantiate Dataset yourself. However, it is useful for optimization:
Memory Efficiency: For massive datasets, convert
Data\(\to\)Datasetand delete the originalData/DataFrameto free up RAM.Performance: If you are running multiple models on the same data, creating a
Datasetonce preventsleaspyfrom repeating the conversion process for everyfit()call.
3. Workflow & Best Practices¶
The Standard Workflow¶
The most common and recommended workflow is straightforward:
[CSV / DataFrame] -> Data.from_dataframe() -> [Data Object] -> model.fit(data)
Inside model.fit(data), Leaspy automatically converts your Data object into a Dataset for computation.
Guidelines for model.fit()¶
Input Type |
Verdict |
Reason |
|---|---|---|
|
✅ Recommended |
Safe & Standard. Handles all model types (including Joint models) correctly. Easy to inspect before fitting. |
|
⚡ Optimization |
Fast. Use for heavy datasets or repeated experiments to skip internal conversion steps. |
|
❌ Avoid |
Risky. Fails for complex models (e.g., |